liger
Note: This package is no longer maintained as I no longer actively use it myself.
For gene set enrichment analysis in R, please check out instead fgsea or clusterProfiler. I hope this repo and its vignettes may still be useful as resource for understanding such gene set enrichment analysis.
Lightweight Iterative Gene set Enrichment in R (LIGER)
Gene Set Enrichment Analysis (GSEA) is a computational method that determines whether an a priori defined set of genes shows statistically significant, concordant differences between two biological states. The original algorithm is detailed in Subramanian, Tamayo, et al. with Java implementations available through the Broad Institute.
The liger package provides a lightweight R implementation of this enrichment test on a list of values. Given a list of values, such as p-values or log-fold changes derived from differential expression analysis or other analyses comparing biological states, this package enables you to test a priori defined set of genes for enrichment to enable interpretability of highly significant or high fold-change genes.
Tutorials
- Gene Set Enrichment Analysis with LIGER
- Interpreting Enrichment Scores and Edge Values
- Exploring Permutation P-values
- Highlighting the power of gene set enrichment analysis using simulation
Sample plots
Testing individual gene sets
> gsea(values=vals, geneset=gs, mc.cores=1, plot=TRUE)

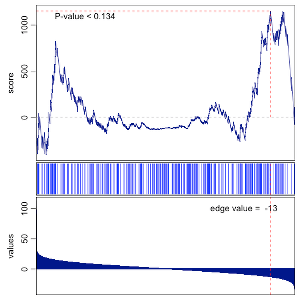
Testing multiple gene sets
> ptm <- proc.time()
> bulk.gsea(vals, org.Hs.GO2Symbol.list[1:10])
p.val q.val sscore edge
GO:0000002 0.00009999 0.0000000 2.6139724 70.912194
GO:0000003 0.25647435 0.4074000 0.5400972 13.170093
GO:0000012 0.24347565 0.4074000 0.5550191 8.392397
GO:0000014 0.13428657 0.3779000 0.6906745 -4.458762
GO:0000018 0.14448555 0.7215200 -0.6498716 11.111976
GO:0000022 0.33446655 0.8023667 -0.5014560 -11.015244
> proc.time() - ptm
user system elapsed
6.200 0.065 6.414
> ptm <- proc.time()
> iterative.bulk.gsea(values=vals, set.list=org.Hs.GO2Symbol.list[1:10])
p.val q.val sscore edge
GO:0000002 0.00009999 0.00000 2.6054741 70.912194
GO:0000003 0.25867413 0.41730 0.5352656 13.170093
GO:0000012 0.24727527 0.41730 0.5545500 8.392397
GO:0000014 0.14128587 0.40300 0.6903051 -4.458762
GO:0000018 0.13988601 0.72684 -0.6485933 11.111976
GO:0000022 0.31666833 0.81440 -0.4975180 -11.015244
> proc.time() - ptm
user system elapsed
11.250 0.107 5.320
Install
# The easiest way to get liger is to install it from CRAN:
install.packages("liger")
# Or get the latest development version from GitHub:
require(devtools)
devtools::install_github("JEFworks/liger")
Contributing
We welcome any bug reports, enhancement requests, and other contributions. To submit a bug report or enhancement request, please use the liger GitHub issues tracker. For more substantial contributions, please fork this repo, push your changes to your fork, and submit a pull request with a good commit message.